Available Reaction Networks
iso7, aprox13, aprox19, and aprox21
These are alpha-chains (with some other nuclei) from Frank Timmes.
These networks share common rates (from Microphysics/rates),
plasma neutrino loses (from Microphysics/neutrinos), and
electron screening (from Microphysics/screening).
Energy generation.
These networks store the total binding energy of the nucleus in MeV as
bion(:). They then compute the mass of each nucleus in grams as:
where \(m_n\), \(m_p\), and \(m_e\) are the neutron, proton, and electron
masses, \(A_k\) and \(Z_k\) are the atomic weight and number, and \(B_k\)
is the binding energy of the nucleus (converted to grams). \(M_k\)
is stored as mion(:) in the network.
The energy release per gram is converted from the rates as:
where \(N_A\) is Avogadro’s number (to convert this to “per gram”) and \(\edotnu\) is the neutrino loss term.
general_null
general_null is a bare interface for a nuclear reaction network –
no reactions are enabled. The
data in the network is defined at compile type by specifying an
inputs file. For example,
networks/general_null/triple_alpha_plus_o.net would describe the
triple-\(\alpha\) reaction converting helium into carbon, as
well as oxygen and iron. This has the form:
# name short name aion zion
helium-4 He4 4.0 2.0
carbon-12 C12 12.0 6.0
oxygen-16 O16 16.0 8.0
iron-56 Fe56 56.0 26.0
The four columns give the long name of the species, the short form that will be used for plotfile variables, and the mass number, \(A\), and proton number, \(Z\).
The name of the inputs file by one of two make variables:
NETWORK_INPUTS: this is simply the name of the “.net” file, without any path. The build system will look for it in the current directory and then in$(MICROPHYSICS_HOME)/networks/general_null/.For the example above, we would set:
NETWORK_INPUTS := triple_alpha_plus_o.net
GENERAL_NET_INPUTS: this is the full path to the file. For example we could set:GENERAL_NET_INPUTS := /path/to/file/triple_alpha_plus_o.net
At compile time, the “.net” file is parsed and a network header
network_properties.H is written using the python script
write_network.py. The make rule for this is contained in
Make.package.
ignition_chamulak
This network was introduced in our paper on convection in white dwarfs as a model of Type Ia supernovae [ZingaleNonakaAlmgren+11]. It models carbon burning in a regime appropriate for a simmering white dwarf, and captures the effects of a much larger network by setting the ash state and energetics to the values suggested in [ChamulakBrownTimmesDupczak08].
Energy generation.
The binding energy, \(q\), in this network is interpolated based on the density. It is stored as the binding energy (ergs/g) per nucleon, with a sign convention that binding energies are negative. The energy generation rate is then:
(this is positive since both \(q\) and \(dY/dt\) are negative)
ignition_reaclib
ignition_simple
This is the original network used in our white dwarf convection studies [ZingaleAlmgrenBell+09]. It includes a single-step \(^{12}\mathrm{C}(^{12}\mathrm{C},\gamma)^{24}\mathrm{Mg}\) reaction. The carbon mass fraction equation appears as
where \(N_A \left <\sigma v\right>\) is evaluated using the reaction rate from (Caughlan and Fowler 1988). The Coulomb screening factor, \(f_\mathrm{Coul}\), is evaluated using the general routine from the Kepler stellar evolution code (Weaver 1978), which implements the work of (Graboske 1973) for weak screening and the work of (Alastuey 1978 and Itoh 1979) for strong screening.
kpp
powerlaw
This is a simple single-step reaction rate. We will consider only two species, fuel, \(f\), and ash, \(a\), through the reaction: \(f + f \rightarrow a + \gamma\). Baryon conservation requires that \(A_f = A_a/2\), and charge conservation requires that \(Z_f = Z_a/2\). We take our reaction rate to be a powerlaw in temperature. The standard way to write this is in terms of the number densities, in which case we have
with
Here, \(r_0\) sets the overall rate, with units of \([\mathrm{cm^3~s^{-1}}]\), \(T_0\) is a reference temperature scale, and \(\nu\) is the temperature exponent, which will play a role in setting the reaction zone thickness. In terms of mass fractions, \(n_f = \rho X_a / (A_a m_u)\), our rate equation is
We define a new rate constant, \(\rt\) with units of \([\mathrm{s^{-1}}]\) as
where \(\rho_0\) is a reference density and \(T_a\) is an activation temperature, and then our mass fraction equation is:
Finally, for the energy generation, we take our reaction to release a specific energy, \([\mathrm{erg~g^{-1}}]\), of \(\qburn\), and our energy source is
There are a number of parameters we use to control the constants in
this network. This is one of the few networks that was designed
to work with gamma_law as the EOS.
rprox
This network contains 10 species, approximating hot CNO, triple-\(\alpha\), and rp-breakout burning up through \(^{56}\mathrm{Ni}\), using the ideas from [WallaceWoosley81], but with modern reaction rates from ReacLib [CAF+10] where available. This network was used for the X-ray burst studies in [MaloneZingaleNonaka+14], [ZingaleMaloneNonaka+15], and more details are contained in those papers.
triple_alpha_plus_cago
This is a 2 reaction network for helium burning, capturing the \(3\)-\(\alpha\) reaction and \(\isotm{C}{12}(\alpha,\gamma)\isotm{O}{16}\). Additionally, \(^{56}\mathrm{Fe}\) is included as an inert species.
subch networks
The networks subch_full and subch_approx recreate an aprox13 alpha-chain + including a bypass rate for \(\isotm{C}{12}(\alpha, \gamma)\isotm{O}{16}\) discussed in [ShenBildsten09]. This is appropriate for explosive He burning.
[ShenBildsten09] discuss the sequences:
\(\isotm{C}{14}(\alpha, \gamma)\isotm{O}{18}(\alpha, \gamma)\isotm{Ne}{22}\) at high temperatures (T > 1 GK). We don’t consider this.
\(\isotm{N}{14}(\alpha, \gamma)\isotm{F}{18}(\alpha, p)\isotm{Ne}{21}\) is the one they consider important, since it produces protons that are then available for \(\isotm{C}{12}(p, \gamma)\isotm{N}{13}(\alpha, p)\isotm{O}{16}\).
This leaves \(\isotm{Ne}{21}\) as an endpoint, which we connect to the other nuclei by including \(\isotm{Na}{22}\).
For the \(\isotm{C}{12} + \isotm{C}{12}\), \(\isotm{C}{12} + \isotm{O}{16}\), and \(\isotm{O}{16} + \isotm{O}{16}\) rates, we also need to include:
\(\isotm{C}{12}(\isotm{C}{12},n)\isotm{Mg}{23}(n,\gamma)\isotm{Mg}{24}\)
\(\isotm{O}{16}(\isotm{O}{16}, n)\isotm{S}{31}(n, \gamma)\isotm{S}{32}\)
\(\isotm{O}{16}(\isotm{C}{12}, n)\isotm{Si}{27}(n, \gamma)\isotm{Si}{28}\)
Since the neutron captures on those intermediate nuclei are so fast, we leave those out and take the forward rate to just be the first rate. We do not include reverse rates for these processes.
subch_full
subch_full does not create an effective rate for \((\alpha, \gamma)\) and \((\alpha, p)(p, \gamma)\) (i.e. combine them assuming proton equilibrium). Therefore, we need to explicitly include the intermediate nuclei produced in the \((\alpha,p)\) reactions. In all, 28 nuclei and 107 rates are included.
This network is generated via pynucastro using the subch_full.py script.
The overall network appears as:
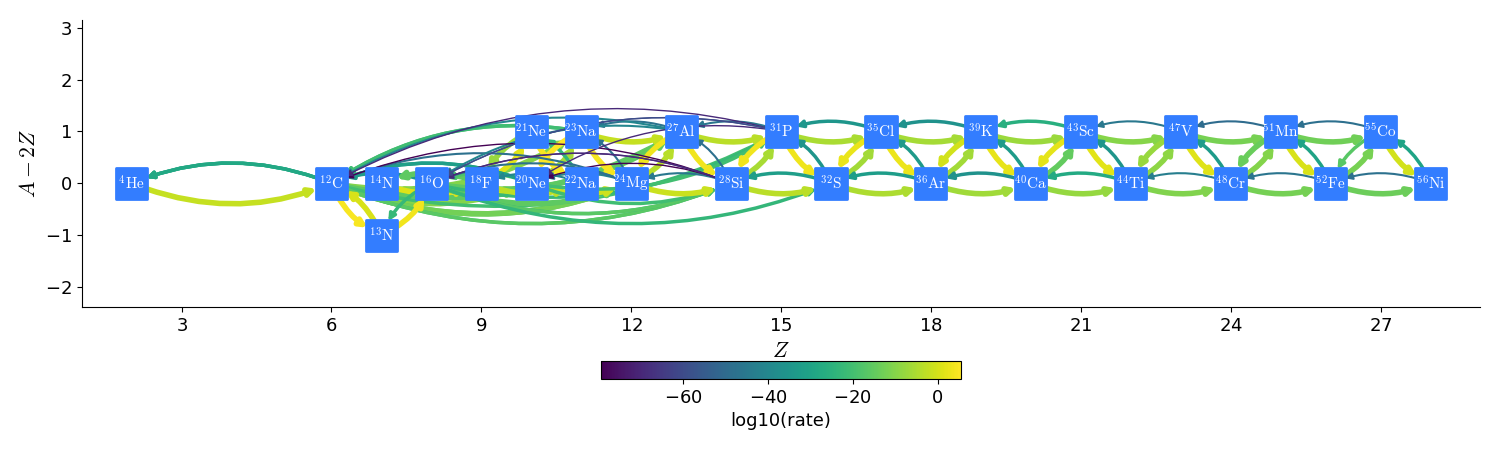
subch_approx
subch_approx approximates subch_full by combining some of the \(A(\alpha,p)X(p,\gamma)B\) links with \(A(\alpha,\gamma)B\), allowing us to drop the intermediate nucleus \(X\). We do this for \(\isotm{Cl}{35}\), \(\isotm{K}{39}\), \(\isotm{Sc}{43}\), \(\isotm{V}{47}\), \(\isotm{Mn}{51}\), and \(\isotm{Co}{55}\). The resulting network appears as:
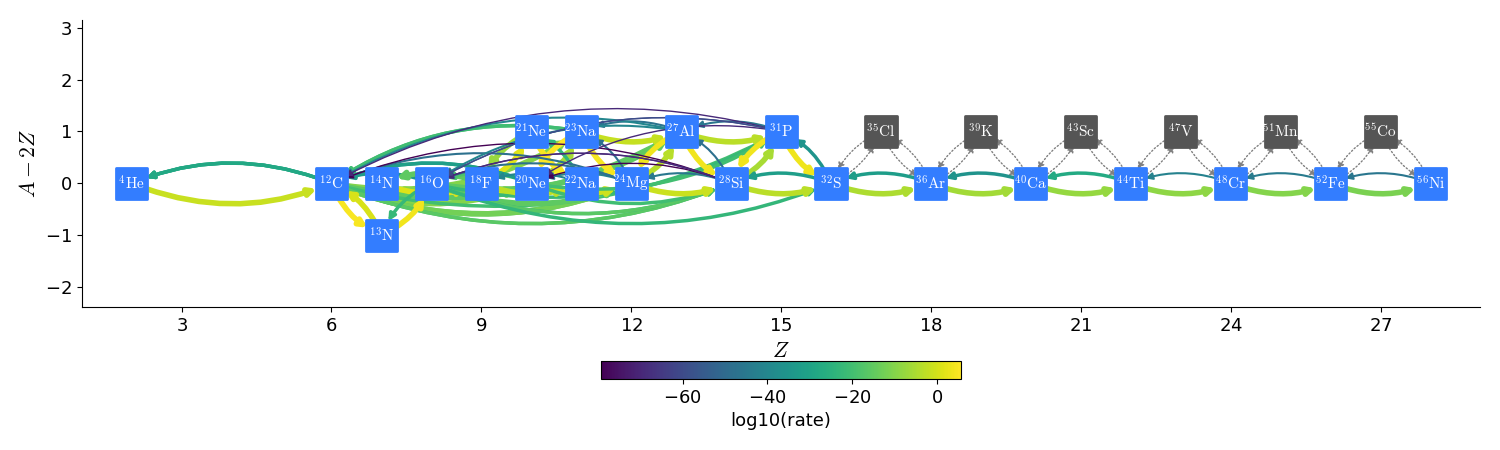
The nuclei in gray are not part of the network, but the links to them are approximated. This reduces the number of nuclei compared to subch_full from 28 to 22.
disabling rates
For both subch_full and subch_approx, there are 2 runtime parameters that can be used to disable rates:
network.disable_p_c12__n13: if set to1, then the rate \(\isotm{C}{12}(p,\gamma)\isotm{N}{13}\) and its inverse are disabled.network.disable_he4_n13__p_o16: if set to1, then the rate \(\isotm{N}{13}(\alpha,p)\isotm{O}{16}\) and its inverse are disabled.
Together, these parameters allow us to turn off the sequence \(\isotm{C}{12}(p,\gamma)\isotm{N}{13}(\alpha, p)\isotm{O}{16}\) that acts as a bypass for \(\isotm{C}{12}(\alpha, \gamma)\isotm{O}{16}\).