Checkpoint Embiggener
Within the Castro distribution, there is the capability to “grow” a
checkpoint file so that a calculation can be restarted in a larger
domain covered by grid cells a factor of two or four coarser than the
existing coarsest level. Instructions for how to do so are in the
Castro/Util/ConvertCheckpoint/README file and are included here.
Upon restart the existing data in the checkpoint file will be used to
fill the region of the previous computational domain, and the new
regions will be filled by some other means, typically interpolation
from a 1D model file.
Star in Corner (star_at_center = 0)
In this section we consider the case where the star (or feature of interest) is centered at the lower left corner of the domain, e.g. you are modeling only one quarter of the star in 2D, or an octant of the star in 3D. Then you only want to grow the domain in the “high side” directions (e.g., to the upper right).
Converting the Checkpoint File
Let’s say you have a checkpoint file, chk00100 with 5 levels of
refinement and a (real) problem domain size \(P\) and (integer)
domain size \(D\) at level 0. The inputs file that created this
might have contained:
max_step = 100
amr.max_level = 5
amr.n_cell = D D
geometry.prob_lo = 0 0
geometry.prob_hi = P P
amr.ref_ratio = 4 4 4 4
Now let’s suppose that you want to grow the domain by a factor of 8 and cover that new larger domain with a level that is a factor of 2 coarser than the existing level 0 grids.
First, set
DIM =in the GNUmakefile, and typemakein theUtil/ConvertCheckpoint/directory. This will make an executable from theEmbiggen.cppcode.Run the embiggening code as follows:
Embiggen2d.Linux.Intel.Intel.ex checkin=chk00100 checkout=newchk00050 ref_ratio=2 grown_factor=8 star_at_center=0
(Your executable may have a slightly different name depending on the compilers you built it with.)
This will create a new checkpoint directory, called
newchk00050, that represents a simulation with one additional level of refinement coarser than the previous level 0 grids by a factor ofref_ratio(in this case, 2). The new domain will be a factor ofgrown_factor(in this case, 8) larger than the previous domain.Note
ref_ratiomust be 2 or 4, because those are the only acceptable values in Castro.grown_factorcan be any reasonable integer; but it’s only been tested with 2, 3, 4 and 8. It does not need to be a multiple of 2.
Restarting from a Grown Checkpoint File
You should now be able to restart your calculation using newchk00050.
Your inputs file should now contain lines like:
max_step = 51
amr.restart = newchk00050
amr.max_level = 6
amr.n_cell = 4D 4D
geometry.prob_lo = 0 0
geometry.prob_hi = 8P 8P
castro.grown_factor = 8
castro.star_at_center = 0
amr.ref_ratio = 2 4 4 4 4
Important:
Unlike earlier, you may now set
amr.max_levelto be at most one greater than before, but you need not set it that high. For example, you could setamr.max_levelthe same as before and you would lose data at the finest refinement level. You may not setamr.max_level= 0, however, because we have no data at the new level 0 until we average down from the new level 1 after the restart.You must set
amr.n_cell= (grown_factor/ref_ratio) \(\times\) (the previous value ofamr.n_cell). In this caseamr.n_cell= (8/2)*D = 4D.You must set
amr.prob_hito be a factor ofgrown_factorgreater than the previous value ofamr.prob_hi.You must insert the value of
ref_ratioused in the Embiggen call as the first value in the list ofamr.ref_ratio, since that will now be the refinement ratio between the new level 0 and the new level 1.You must set
castro.grown_factorin your inputs file equal to the value ofgrown_factoryou used when you called Embiggen*ex so that Castro knows how big the original domain was.Note that if you have run 100 steps at the original level 0, that would be equivalent to 50 steps at the new level 0 because you coarsened by a factor of 2. Thus once you re-start from the new checkpoint directory, the next step will be 51, not 101. Make sure to keep track of your plotfiles accordingly.
Don’t forget to adjust
max_denerr_levand comparable variables to control the number of fine levels you now want. If you want to have 6 levels of refinement after restart, then make suremax_denerr_lev, etc, are set high enough. If you only want to have 5 levels of refinement (where the new level 5 would now be a factor ofref_ratiocoarser than the previous level 5), make sure to adjustmax_denerr_levaccordingly as well.
Star at Center of Domain (star_at_center = 1)
Now let’s assume that the star (or feature of interest) is centered at the center of the domain in 2D or 3D Cartesian coordinates. We will later consider the case of 2D cylindrical (r-z) coordinates in which the star is centered at the left midpoint.
Converting the Checkpoint File
Suppose that you want to grow the domain by a factor of 2 and cover that new larger domain with a level that is a factor of 2 coarser than the existing level 0 grids.
After you build the Embiggen executable, you type:
Embiggen2d.Linux.Intel.Intel.ex checkin=chk00100 checkout=newchk00050 ref_ratio=2 grown_factor=2 star_at_center=1
Note that
ref_ratiomust still be 2 or 4grown_factorcan only be 2 or 3 in this case.
Restarting from a Grown Checkpoint File
Your inputs file for restarting would now look like:
max_step = 51
amr.restart = newchk00050
amr.max_level = 6
amr.n_cell = D D
geometry.prob_lo = -P/2 -P/2
geometry.prob_hi = 3P/2 3P/2
castro.grown_factor = 2
castro.star_at_center = 1
amr.ref_ratio = 2 4 4 4 4
Cylindrical Coordinates
In the case of 2D cylindrical (r-z) coordinates in which the star is
centered at the left edge but vertical midpoint of the domain, the
embiggening procedure is the same as above (with star_at_center =
1) but the inputs file for restart is slightly different in that
geometry.prob_lo is modified in the z- but not the r-direction. If
we consider the original inputs file to look like:
max_step = 100
amr.max_level = 6
amr.n_cell = D 2D
geometry.prob_lo = 0 0
geometry.prob_hi = P 2P
amr.ref_ratio = 4 4 4 4
then an inputs file for restart would look like:
amr.restart = newchk00050
amr.max_level = 6
amr.n_cell = D 2D
geometry.prob_lo = 0 -P
geometry.prob_hi = 2P 3P
castro.grown_factor = 2
castro.star_at_center = 1
amr.ref_ratio = 2 4 4 4 4
Some results:
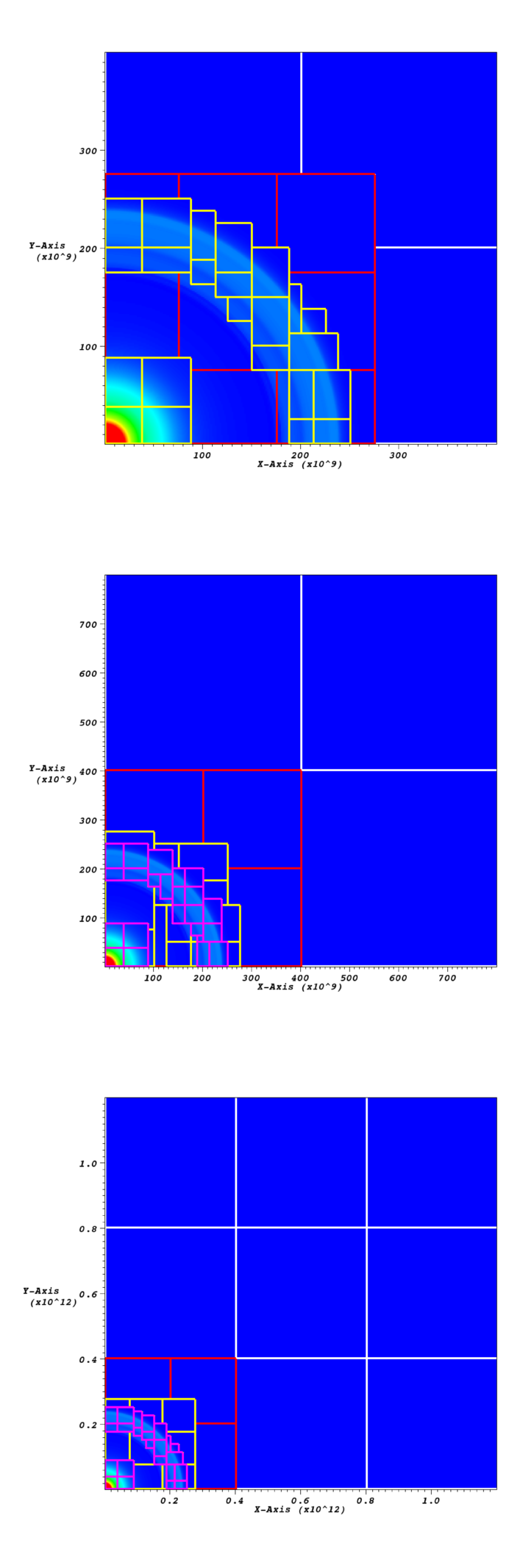
Fig. 8 Data from checkpoint file before and after the domain has been
coarsened and grown. This case uses star_at_center = 0 and
ref_ratio = 2. The first grown example has grown_factor =
2, the second has grown_factor = 3. In all figures the level 0
grids are shown in white, the level 1 grids in red, the level 2
grids in yellow, and in the grown figures, the level 3 grids are in
pink.
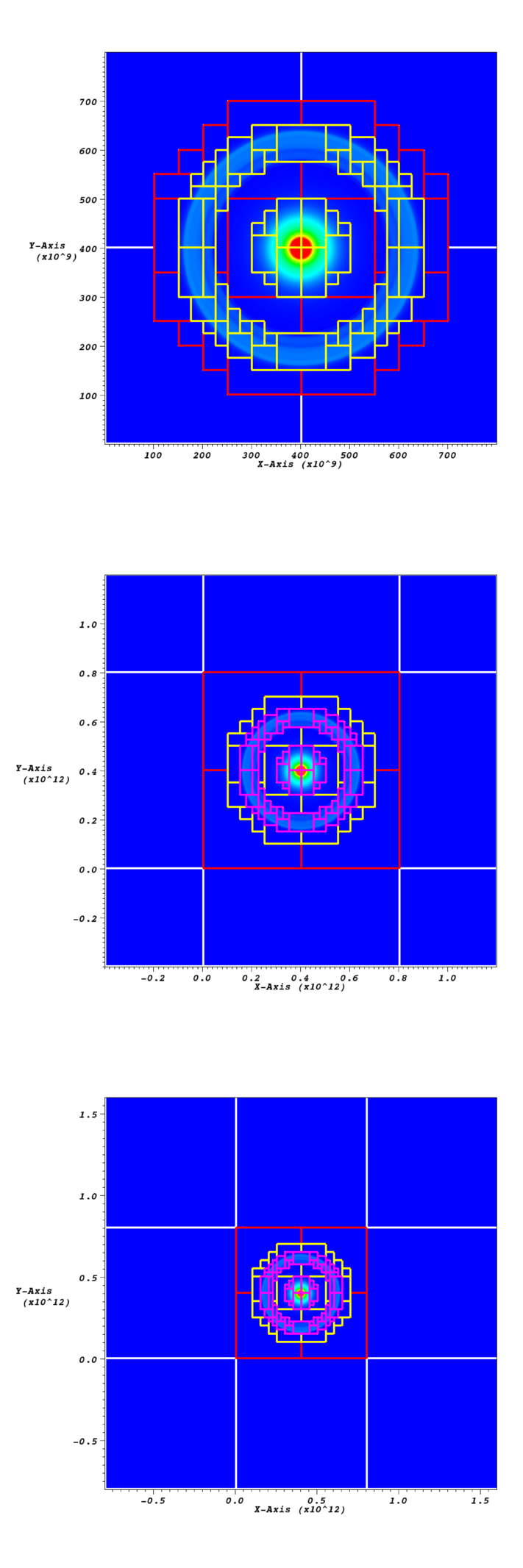
Fig. 9 Data from checkpoint file before and after the domain has been
coarsened and grown. This case uses star_at_center = 1 and
ref_ratio = 2. The first grown example has grown_factor =
2, the second has grown_factor = 3. In all figures the level 0
grids are shown in white, the level 1 grids in red, the level 2
grids in yellow, and in the grown figures, the level 3 grids are in
pink.