Visualization
Plotfiles
MAESTROeX outputs plotfiles specifically for visualization and analysis. The table below lists the quantities stored in a plotfile. Not all of these may be present, dependent on what options were used in creating the plotfile.
By default, plotfiles store double precision data, but if:
fab.format = NATIVE_32
is set in the inputs file, then the data is converted to single precision before outputting—this is done to reduce file sizes.
plotfile variable name |
description |
runtime parameter controlling output |
|---|---|---|
x_vel |
\(\ut\) |
– |
y_vel |
\(\vt\) |
– |
z_vel |
\(\wt\) |
3-d runs only |
density |
\(\rho\) |
– |
rhoh |
\((\rho h)\) |
use_tfromp = F or (use_tfromp = T and plot_h_with_use_tfromp = T) |
h |
\((\rho h)/\rho\) |
use_tfromp = F or (use_tfromp = T and plot_h_with_use_tfromp = T) |
X(*) |
\((\rho X_k)/\rho\) |
plot_spec |
tracer |
tracers |
plot_trac |
w0_x |
\(w_0 \er \cdot \ex\) |
plot_base |
w0_y |
\(w_0 \er \cdot \ey\) |
plot_base |
w0_z |
\(w_0 \er \cdot \ez\) |
plot_base |
divw0 |
\(\nabla \cdot w_0\) |
plot_base |
rho0 |
\(\rho_0\) |
plot_base |
rhoh0 |
\((\rho h)_0\) |
plot_base |
h0 |
\((\rho h)_0/\rho_0\) |
plot_base |
p0 |
\(p_0\) |
plot_base |
radial_velocity |
\(\Ubt \cdot \er + w_0\) |
spherical == 1 |
circum_velocity |
\(|\Ubt - (\Ubt \cdot \er) \er |\) |
spherical == 1 |
magvel |
\(| \Ubt + w_0 \er |\) |
|
momentum |
\(\rho | \Ubt + w_0 \er |\) |
|
vort |
\(| \nabla \times \Ubt |\) |
|
S |
\(S\) |
– |
rhopert |
\(\rho - \rho_0\) |
– |
rhohpert |
\((\rho h) - (\rho h)_0\) |
use_tfromp = F or (use_tfromp = T and plot_h_with_use_tfromp = T) |
tfromp |
\(T(\rho, p_0, X_k)\) |
– |
tfromh |
\(T(\rho, h, X_k)\) |
use_tfromp = F or (use_tfromp = T and plot_h_with_use_tfromp = T) |
deltaT |
\([T(\rho, h, X_k) -\) \(T(\rho, p_0, X_k)]\) \(/T(\rho, h, X_k)\) |
use_tfromp = F or (use_tfromp = T and plot_h_with_use_tfromp = T) |
deltap |
\(|p(\rho,h,X_k)-p_0|/p_0\) |
use_tfromp = F or (use_tfromp = T and plot_h_with_use_tfromp = T) |
tpert |
\(T(\rho,h,X_k)-\overline{T}\) if use_tfromp = F; \(T(\rho,p_0,X_k)-\overline{T}\) otherwise |
– |
Machnumber |
\(|\Ubt+w_0\er |/c(\rho,p_0,X_k)\) |
– |
soundspeed |
\(c(\rho,p_0,X_k)\) |
plot_cs |
deltagamma |
\(\Gamma_1-\gammabar\) |
– |
entropy |
\(s(\rho,p_0,X_k)\) |
– |
entropypert |
\([s(\rho,p_0,X_k) -\) \(\overline{s}]/\overline{s}\) |
– |
sponge or sponge_fdamp |
\(1/(1+\Delta t\kappa \fdamp)\) by default; \(\fdamp\) if plot_sponge_fdamp = T |
– |
pi |
\(\pi\) |
– |
gpi_x |
\(\nabla \pi \cdot \ex\) |
plot_gpi |
gpi_y |
\(\nabla \pi \cdot \ey\) |
plot_gpi |
gpi_z |
\(\nabla \pi \cdot \ez\) |
plot_gpi |
pioverp0 |
\(\pi / p_0\) |
plot_base |
p0pluspi |
\(p_0 + \pi\) |
plot_base |
omegadot(*) |
\(\dot{\omega}_k\) |
plot_omegadot |
enucdot |
\((\rho \Hnuc)/\rho\) |
plot_Hnuc |
Hext |
\((\rho \Hext)/\rho\) |
plot_Hext |
eta_rho |
\(\etarho\) |
plot_eta |
thermal |
\(\nabla \cdot \kappa\nabla T\) |
use_thermal_diffusion |
conductivity |
\(\kappa(\rho, T,X_k)\) |
use_thermal_diffusion |
ad_excess |
\(\nabla - \nabla_\mathrm{ad}\) |
plot_ad_excess |
particle_count |
number of particles in a cell |
use_particles |
processor_number |
processor number containing the cell’s data |
plot_processors |
pi_divu |
\(\pi \nabla \cdot\tilde{\Ub}\) (a measure of energy conservation) |
plot_pidivu |
Small vs. regular plotfiles
MAESTROeX can manage two independent sets of plotfiles. This allows you to output the default plotfiles, which contain a lot of variables, sparsely, and output small-plotfiles much more frequently. A small-plotfile is controlled by the analogous runtime parameters as the main plotfiles:
small_plot_intis the interval in steps between successive plotfilessmall_plot_deltatis the interval in time between successive plotfilessmall_plot_base_nameis the base name that prefixes the plotfiles. The default is smallplt
The fields that are stored in the small plotfiles is set by the runtime
parameter small_plot_vars. This should be a (space-separated) list of the
parameter names to be included in the plot file.
Visualizing with Amrvis
Once the code is built, you visualize a dataset as:
amrvis3d.Linux.g++.gfortran.ex pltfile
where pltfile is the name of the plotfile directory. Different variables can be selected from the drop down menu at the top. Middle and right clicking in 3-d select the slice planes, and shift + middle or right will extract 1-d lines through the data. In 2-d, middle and right clicking alone extract 1-d lines.
If Amrvis cannot find the Palette file, then the plots will be in grayscale. To fix this, copy the amrvis.defaults and Palette files to your home directory and edit amrvis.defaults so that the palette line points to the Palette file, e.g.:
palette /home/username/Palette
Visualizing with VisIt
VisIt recognizes MAESTROeX plotfiles as being in the BoxLib format.
Visualizing with yt
yt is a Python package for analyzing and visualizing simulation data, and understand that AMReX data from MAESTROeX and CASTRO (along with many other simulation codes). For more information, see the yt homepage at http://yt-project.org/ and [TurkSmithOishi+11].
Some sample scripts that use yt with MAESTROeX data are contained in
MAESTROeX/Util/yt/.
plotsinglevar.py
plotsinglevar.py does visualizations of 2-d AMReX plotfiles,
and slices through 3-d AMReX plotfiles. A simple plot can be made
via:
python plotsinglevar.py --log -o test.png plt00000/ tfromp
This will make a plot of “tfromp” from the plotfile plt00000 with log scaling,
and store the output in test.png, as showing the figure below.
If you don’t do ‘-o’, then a default output filename consisting of the
plotfile name + component will be used.
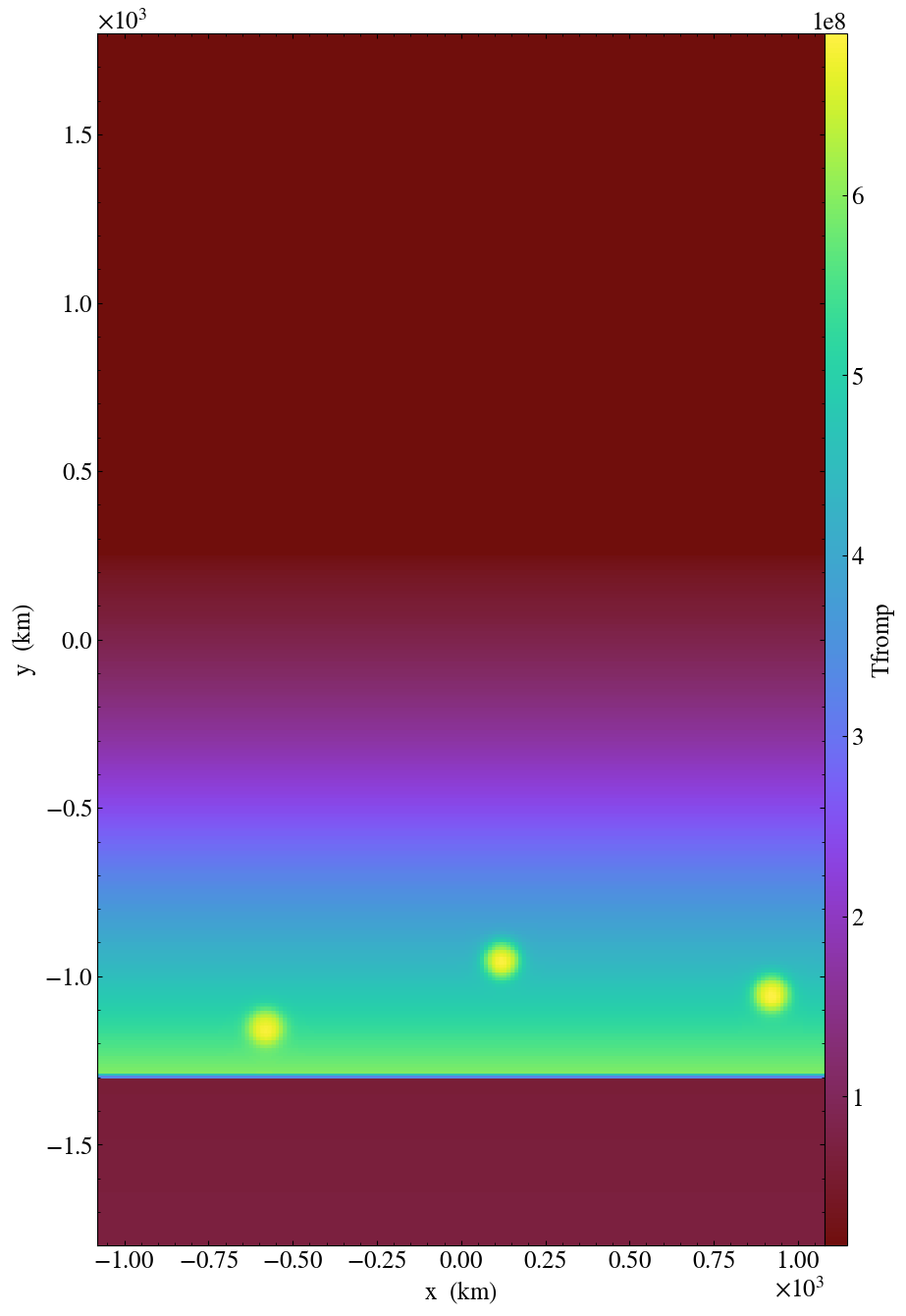
Fig. 14 Plot of reacting_bubble done with the python
script plotsinglevar.py.
If you list 2 different variables after the plotfile name, then they will be plotted side-by-side in a single figure. For example,
python plotsinglevar.py plt00000/ tfromp Hnuc
produces the output shown below:
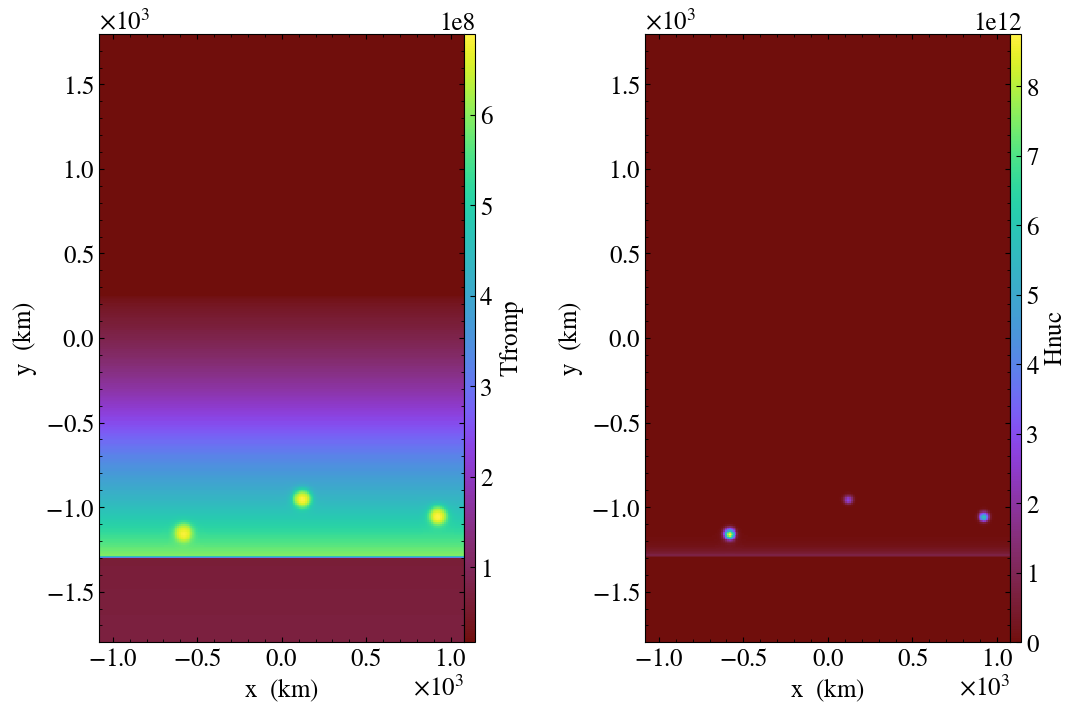
Fig. 15 Plot of reacting_bubble done with the python script
plotsinglevar.py showing 2 variables plotted from a single
plotfile.
Additional options include ‘-min’ to specify the minimum data
value, and ‘-max’ to specify the maximum data value. Running the script with
the flag -h will list the available options.
3-d support is available. When run as with a plotfile name and variable, it will plot slices (\(x\)-\(y\), \(x\)-\(z\), and \(y\)-\(z\)) through the center of the domain.
contourcompare.py
contourcompare.py takes two or three plotfiles and a single variable as arguments
and plots contours of the datasets on the same set of axes. This is
facilitates comparisons of different runs. Running the script with the flag -h
will give the full list of available options.
For example:
python contourcompare.py tfromp plt00000 other_plt00000
will make a contour plot of the variable tfromp from the data in
plt00000 and other_plt00000 shown on the same axes.
runtimevis.py
The runtimevis.py script is designed to be run from a submission
script to produce plots from plotfiles as they are produced.
The script itself reads in an inputs file, vis.in, that
describes the variables to plot. From 1 to 6 variables can be
plotting from a plotfile. The script does its best to organize them
in columns and rows to maximize the plot area. The image is always
output at 1280\(\times\)720 pixels, corresponding to 720p HD resolution.
For each variable, a block of the form:
[varname]
min = 1
max = 2
log = 1
is supplied. If min or max are omitted, then the data
limits are computed automatically. If log is omitted, then no
log is taken of the data before plotting. The script is then run as:
python runtimevis.py plt00000